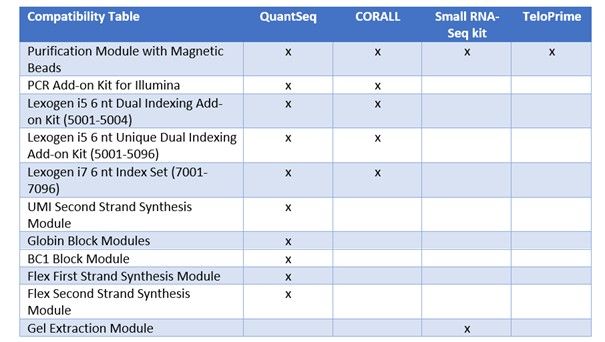
12nt Unique Dual Index System (UDI) for RNA-seq
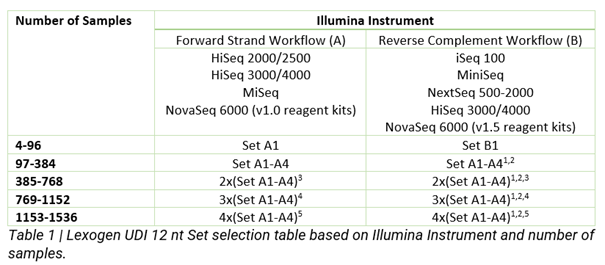
The 12nt UDI sets are an add-on, introduced at the PCR step of Library Preparation, enabling up to 384 unique combinations for higher multiplexing of sequencing libraries on Illumina Flow cells. The Indices are designed to maximize inter-index distance for different sample numbers and index read-out lengths. Read mis-assignment due to index hopping is avoided and index sequence errors can be corrected with the highest accuracy. This system provides the optimized indexing solution for current and future barcoding requirements.

Read mis-assignment caused by Index Hopping can be avoided by using Unique Dual Indexing (UDI). Reads with hopped indices are irreversibly discarded (C). Reads with random Index Sequence Errors resulting in an index not present in the pool are classified undetermined. Accurate error correction can rescue most of these reads making them available for downstream data analysis (B). The percentage values were derived from an RNA-Seq experiment pooling 96 libraries with Lexogen’s 12 nt UDIs and full 12 nucleotide index read-out on an Illumina NextSeq500.
Scalable Index Read-out Length
The design enables scalable read-out lengths of 12, 10 and 8 nucleotides. In ths way, the UDIs support all kinds of requirements for multiplexing, which depend on experiment type, sequencing equipment, desired read depth, and/or the number of pooled libraries. For small sample sizes, short indices (8 or 10nt) are sufficient to ensure high accuracy and reliable error correction. For more than 96 samples however, 8nt index read-out does not allow reliable error correction anymore, and 10 or 12nt read-outs are required.
While needing slightly more sequencing cycles, 12nt index sequences provide the ability to correct up to 3 Index Sequence Errors. Adjustable index read-out length allows tuning your index needs to the experiment design, without the need to purchase separate indexing sets.
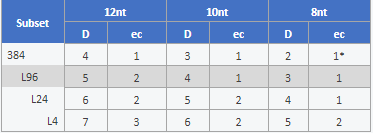
table 1: Inter-index distance (D) and number of errors that can be corrected (ec) are compared for subsets of 384, 96, 24, and 4 libraries and the three possible read-out lengths. For smaller subsets (up to 96 samples) a read-out of 8 or 10 nt allows correction of one error and thus recovery of additional reads. Larger subsets require a read-out of 10 or 12 nt to benefit from the error correction. The ec values represent the number of all errors (including substitutions, insertions, and deletions) that can be confidently corrected, except for *. In this case error correction can only address substitutions.