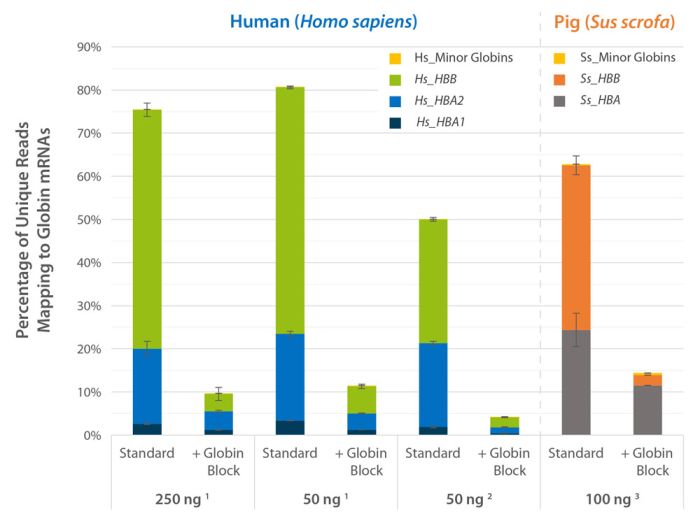
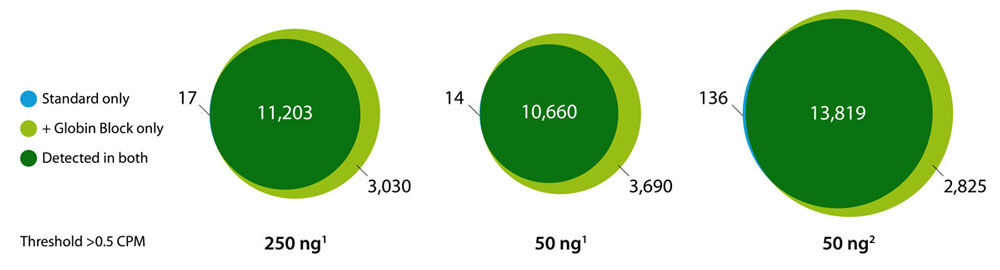
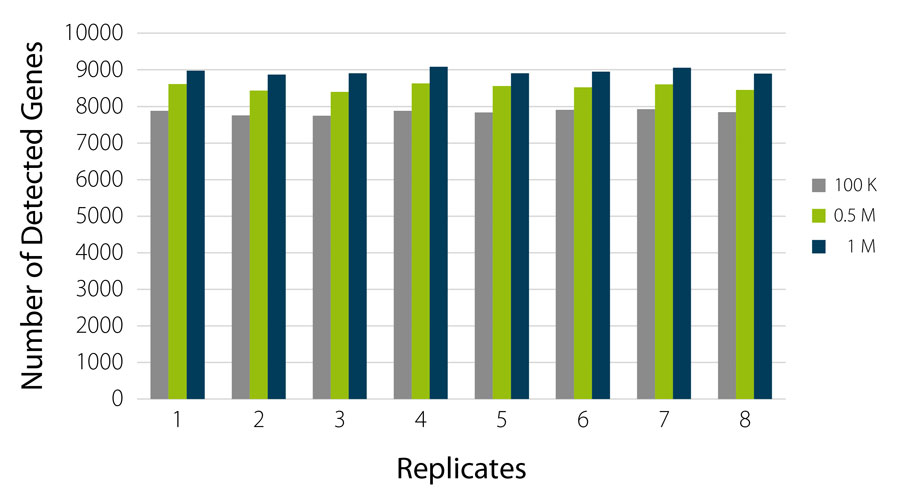
Direct counting for Gene Expression Quantification
No length normalization is required because only one fragment per transcript is produced, enabling accurate determination of gene expression values and making it the best alternative to microarrays and conventional RNA-Seq in gene expression and eQTL studies.
An RNA-Seq Experiment Tailored to your needs
Quant-Seq Modular Kits give the needed flexibility to enable implementation of RNA-seq as part of the experimental pipeline. One Fragment per transcript produced and the use of custom primers makes this kit a perfect solution for cost-saving high-throughput gene expression analysis. Quant-Seq Flex is suitable for targeted sequencing of gene panels, enrichment, detection of fusion genes and various other types of transcripts.
Rapid Turnaround
The straightforward protocol allows generating sequencing-ready libraries in only 4.5 hours, including a hands-on time of less than 2 hours.
Cost Saving Multiplexing
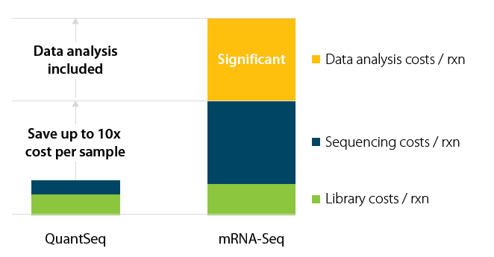
QuantSeq Libraries are intended for a high degree of multiplexing. When combining the 96 i7 indices, included in the kit, with the separately available 96 i5 indices, up to 9.216 samples can be combined on a single lane of an Illumina Flow Cell. This allows substantial sequencing cost saving, as the length restriction in QuantSeq saves sequencing space.
Contact us for prices of more information