Applied Natural Sciences from Fontys University

The research group of Applied Natural Sciences from Fontys University (The Netherlands) focuses on five research fields: Thin Films and Functional Materials (Dr. Jan Bernards), Solar Fuels (Dr. Peter Thüne), Detection and Measurement (Dr. Urs Wyder), Polymers (Ing. Guido Smets) and Life Sciences (Dr. Joost Schoeber and Dr. Anne Loonen).
The Life Science group mainly focuses on the development, optimization, and validation of molecular diagnostic assays to improve health. Currently, the group is working on developing innovative molecular assays to screen for sexually transmitted diseases in patients and wastewater (mirror of society).
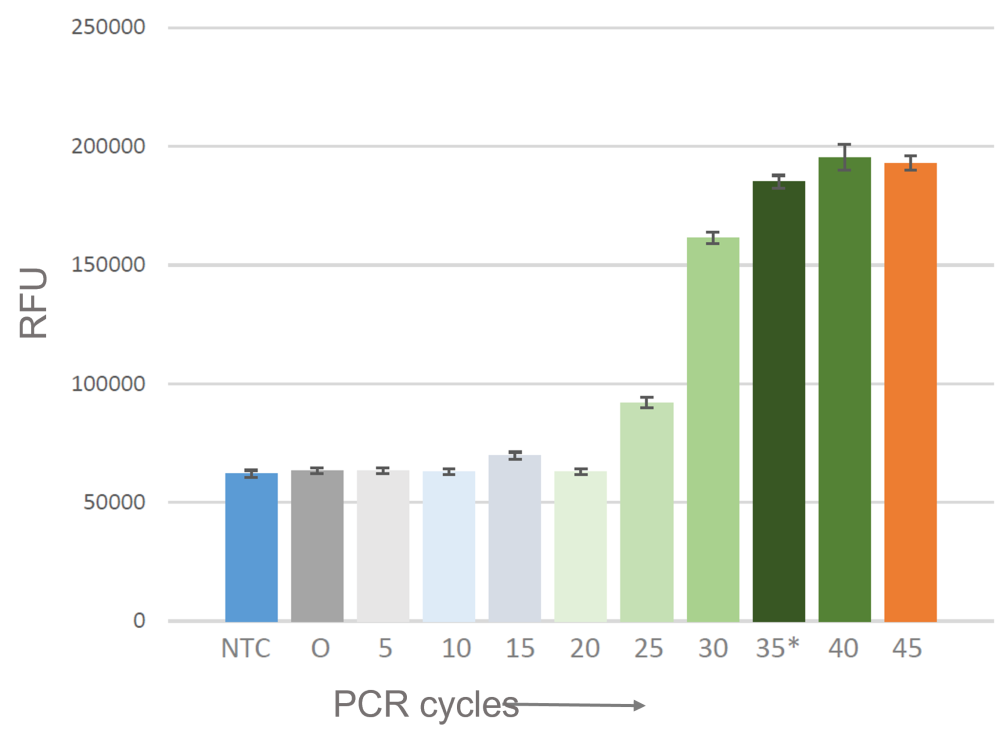
Anouk Poppelaars is doing her bachelor’s thesis at the Life Science department and her main goal is to develop an ultrafast next-generation PCR protocol for the detection of Chlamydia trachomatis (CT). To show her research results, she presented a poster at the BAMA Symposium 2022, organized by the Microbes in Health and Diseases (MHD), University of Groningen. Anouk won three different Prizes: A poster award, and jury and audience prizes.