The SLAMseq kits portfolio includes Explorer and Kinetics Modules, which cover the entire metabolic RNA labelling experiment, from optimizing labelling conditions, to labelling of newly synthesized or existing RNA and alkylation for downstream NGS library prep.
Combining SLAMseq with QuantSeq 3’ mRNA-seq or QuantSeq Flex Targeted RNA-Seq kits provides a complete user-friendly solution for high throughput metabolic RNA-seq experiments.
Total RNA from SLAMseq experiments can be used directly in the QuantSeq protocol. Simply isolate total RNA, alkylate with iodoacetamide, prepare libraries and sequence.
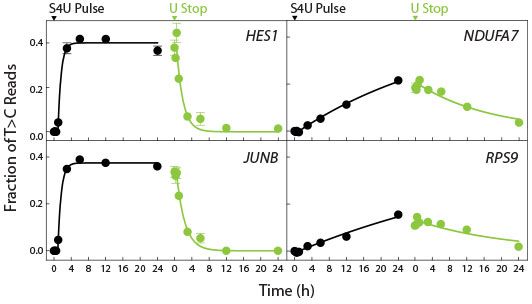
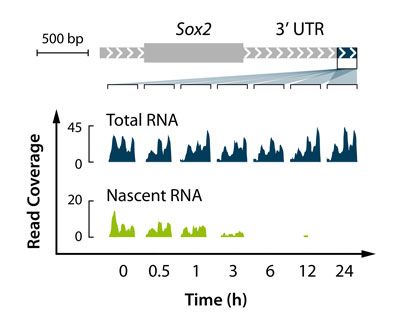
- Analyse transcriptomics-wide kinetics of RNA synthesis and turnover
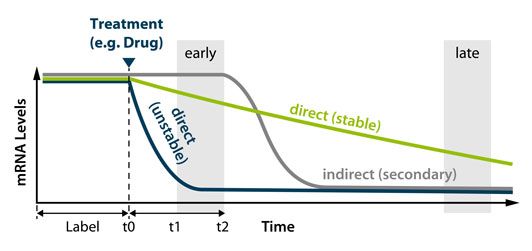
- Measure nascent RNA expression and transcript stability
- Enhance the temporal resolution of differential expression
- Identify primary and secondary transcriptional targets
- No pull-down or biochemical isolation required
- Combine with QuantSeq 3’ mRNA-seq for cost-effective high throughput metabolic sequencing